Abstract
Castleman disease (CD) is a rare lymphoproliferative disorder, that can transform into malignant lymphomas during clinical progress. Histologically CD can be subdivided into a hyaline-vascular variant (CD-HVV), a plasma cell variant (CD-PCV) and an intermediate variant (CD-IV) and further sub-classified into localized unicentric CD (U-CD), and multicentric CD (M-CD). An association with human herpes virus 8 (HHV8) and human immunodeficiency associated virus (HIV) in some CD cases is well documented, though the pathogenesis of CD still remains to be understood (Casper et al. Br J Haematol 2005). In recent years, cytogenetic and molecular studies have demonstrated clonal abnormalities in CD (Fajgenbaum et al. Blood 2017) as well as somatic mutations in follicular dendritic cell sarcoma (FDCS) - a rare clinically aggressive mesenchymal tumor associated with CD-HVV (Griffin et al. Mod Pathol 2015). However, an understanding of the genetic and epigenetic landscape of these rare diseases is lacking. To the best of our knowledge, our study is the first to analyze the genetic and epigenetic features of CD and FDCS associated with CD.
We analyzed 18 cases of CD and 3 cases of FDCS associated with CD by whole exome and/or targeted next generation deep sequencing (NGS). Hybrid-capture NGS [FoundationOne (F1), Foundation Medicine Inc. (FMI), Cambridge, MA] of 343 genes was performed on our 18 CD cases as previously described (Lee et al. Cancer Discov 2016). 100 base-pair paired end whole exome sequencing was performed on our 3 FDCS cases using Sureselect whole exome V5 kit (Agilent Technologies) at an average depth of coverage of 180-fold. Alignment was performed using BWA-MEM with further analysis performed using GATK (version 3), Varscan2, and SNNPET (Agilent Technologies). Annotation of variants was performed using Surecall (version 3.5.1.46, Agilent Technologies) and Seattleseq (University of Washington, Seattle WA; Version 9.10). Variants were additionally curated using Exome Aggregation Consortium (EXAC), Clinvar, 1000 genomes, and Exome Variant Server (EVS). Copy number variation (CNV) analysis was performed with Surecall and CNVSeq, both run with default parameters, and analysis was performed on the "hits" file.
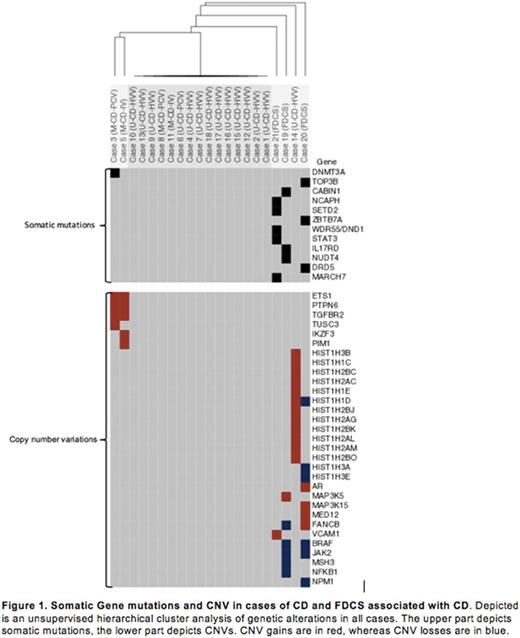
Of our 18 CD cases, 13 were CD-HVV, 3 were CD-PCV and 2 were CD-IV. 14 cases were unicentric and 4 were multicentric. All cases were negative for HHV8 and HIV. A single case of multicentric CD, CD-PVC, showed a somatic DNMT3A L295Q mutation; a gene frequently mutated in hematopoietic neoplasms and possibly related to clonal hematopoiesis of indeterminant potential (Zhang et al. Biomark Res 2017; Yang et al. Nat Rev Cancer 2015). Two other cases of multicentric CD showed common amplification of PTPN6 , ETS1 and TGFBR2 . These genes are known to interface in oncogenesis or critical biologic processes: deletion of PTPN6 results in alteration in dendritic cell function and autoimmunity (Clare et et al. Nano 2013), overexpression of ETS1 is seen in other instances such as breast carcinoma, and leukemia (Dittmer et al. Mol Cancer 2013) and relatively increased expression of TGFBR2 is seen in stromal/vascular tissues. Only one CD-HVV case, presenting clinically untypically aggressive, showed amplifications of HIST genes. These findings could be consistent with a clonal, neoplastic nature to at least a subset of CD cases.
To understand the possible association between CD-HVV and FDCS we also analyzed the mutational landscape of FDCS associated with CD. Our FDCS cases had 3-5 somatic mutations and CNVs in oncogenes and genes known to be critical in tumorigenesis, such as: ZBTB7a - playing a key role in the development from lymphoid progenitors into B-cell lineage, and STAT3 - a transcription factor mutated in NK, T-cell and B-cell disorders (Yu et al. Nat Publ Gr 2014). Given the diverse clinical presentation of FDCS it is not surprising, that previously published mutations seen in FDCS not arising from CD were missing in our study. However, previously described CNVs in FDCS such as NFKBI , and JAK2 were present, next to overlapping CNVs in BRAF and FANCB in two of our cases. In one of our FDCS cases loss of several HISTH genes were further seen. No Translocations or recurrent mutations were seen.
Mughal: Foundation Medicine, Inc: Employment, Other: Stock. Ali: Foundation Medicine, Inc: Employment, Other: Stock.
Author notes
Asterisk with author names denotes non-ASH members.